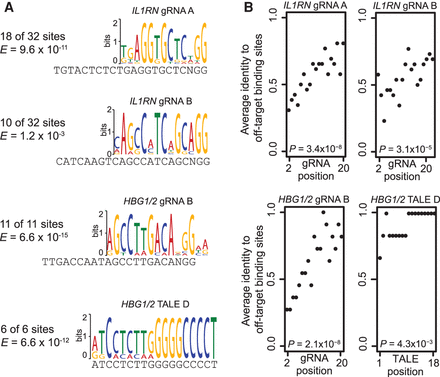
Characterization of dCas9-VP64 and TALE-VP64 off-target binding sites. (A) De novo motif detection was used to identify gRNAs and TALEs responsible for identified off-target binding sites. For dCas9-VP64 targeted with gRNAs, motifs matching two of the IL1RN gRNAs and one of the HBG1/2 gRNAs were identified in the respective binding sites identified with ChIP-seq. For TALE-VP64, no motifs matching the IL1RN TALEs were identified, and one motif matching a HBG1/2 TALE-VP64 was identified. (B) For each off-target binding site identified by ChIP-seq, we performed an unbiased search for sequences that resemble the intended target sequences of each of the gRNAs or TALE identified in A. For this analysis, we considered every possible binding sequence in the called ChIP-seq peak. For dCas9-VP64, we required each possible binding sequence to be followed by the “NGG” PAM sequence. For TALE-VP64, every position in the called binding site was used. Next, for each of the three gRNAs or TALEs identified in A, we aligned the intended target sequence to that of every possible binding sequence, and the sequence with the most matching nucleotides in each binding site was retained. DNA sequence similarity to the target sequence at the matched sites was then plotted as a function of the position in the target sequences. For the three gRNAs investigated, a statistically significant trend toward more similarity at the 3′ end of the gRNA sequence was identified, indicating that the 3′ end of the gRNA is more influential in guiding dCas9-VP64 binding. In contrast, the weak 3′ trend observed for TALE D binding is likely an artifact of low sequence complexity in the 3′ end of the target sequence.











