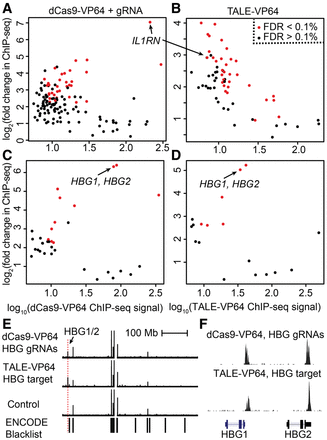
Genome-wide specificity of dCas9-VP64 and TALE-VP64 DNA-binding. ChIP-seq was used to map the genomic locations of dCas9-VP64 targeted to the IL1RN promoter (A), TALE-VP64 targeted to the IL1RN promoter (B), dCas9-VP64 targeted to the HBG1/2 promoters (C), or TALE-VP64 targeted to the HBG1/2 promoters (D). In each plot, points are binding sites that are reproducible in at least two of three replicates. The x-axes are the mean ChIP-seq signal, and the y-axes are the fold-change in signal in samples transfected with dCas9-VP64 or TALE-VP64 transcription factors compared to controls. Red points represent binding sites with a statistically significant increase in signal strength according to analysis with DESeq (false discovery rate <0.1%). (E) As an example of the genome-wide binding specificity dCas9-VP64 and TALE-VP64 targeting, ChIP-seq signal from experiments targeting HBG1/2 was plotted across Chromosome 11. ChIP-seq signals found in both experimental conditions and in the control condition are also found in the ENCODE blacklist, indicating that they are technical artifacts of ChIP-seq and not binding events. Meanwhile, strong ChIP-seq signal was found at the HBG1/2 promoters in the dCas9-VP64 and TALE-VP64 conditions but not in the control condition. (F) The dCas9-VP64 and TALE-VP64 ChIP-seq peaks localize to the intended HBG1/2 promoters.











