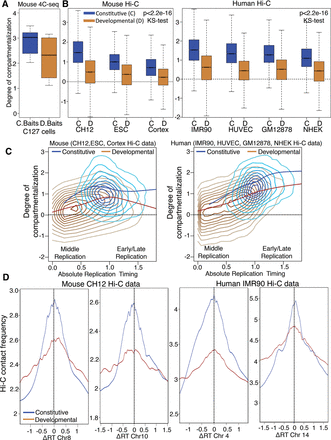
Developmental domains are less compartmentalized than constitutive domains. (A) Degree of compartmentalization (Methods) measured for constitutive baits (C baits), in blue, vs. developmental baits (D baits), in brown. The difference in degree of compartmentalization between C baits and D baits did not have statistical significance (P = 0.7374, KS test); therefore, we performed genome-wide comparison using Hi-C. (B) Degree of compartmentalization for constitutive (C) regions vs. developmental domains (D) (50-kb windows) across three mouse cells types and four human cell types using Hi-C data. The difference in degree of compartmentalization between constitutive domains and developmental domains showed high statistical significance (P < 2.2 × 10−16, KS test). (C) Density contour plot (lighter colors indicate less density) with local regression fitting (loess) for degree of compartmentalization vs. absolute replication timing value for developmental (brown) vs. constitutive (blue) regions. (D) Plot comparing frequency of Hi-C contacts vs. difference in replication timing value for pairs of 50-kb windows using Hi-C data. The top 50 percent of contacts from the Hi-C data was used for the analysis. Blue and brown lines show local regression fitting of the data for constitutive and developmental regions.











