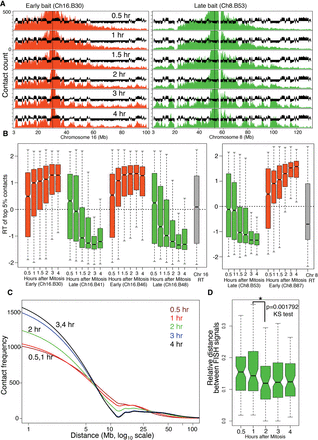
Interphase chromatin compartments are established coincident with TDP. (A) Representative 4C contact count plots for baits positioned in early replicating (red) and late replicating (green) domains for the entire chromosome. 4C profiles are overlaid with C127 replication timing data (black). (B) Distribution of replication timing values for the strongest (top 5% highest z-scores) contacts with early replicating (red) and late replicating (green) baits across Chr 8 and Chr 16. For reference, the RT distributions of the whole chromosomes 8 and 16 are shown in gray. Contacts before the TDP did not have statistical significance, whereas significant contacts begin to appear coincident with the TDP and become more compartmentalized after the TDP (Supplemental Fig. 7A). (C) Loess (local polynomial regression fitting) fitted decay of 4C contact frequency with distance from the bait (decay curve) averaged for all baits from Chr 8 and Chr 16. (D) FISH-based measurement of 3D proximity normalized to the size of the nucleus (relative distance) (Methods) for two late loci (Supplemental Fig. 4) at different points after release from mitosis (nocodazole-free synchrony). There is a significant increase in 3D proximity (P < 0.001792) between before 1 h and after 2 h, which is coincident with the TDP. N = 91, 82, 57, 184, 67 for 30 min, 1, 2, 3 and 4 h, respectively. Mitosis is not included in the time course due to the inability to normalize to the state of condensation of mitotic chromatin (see Supplemental Methods).











