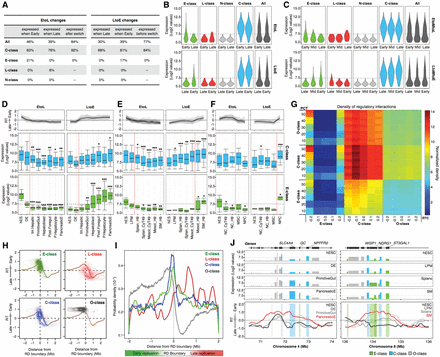
Gene expression regulation and RT changes. (A) RT changes were collected from all transition stages in all differentiation pathways, and the percentages of expressed genes in each stage (before and after the RT switch) were obtained. The first columns in EtoL and LtoE changes contain the percentages of genes expressed calculated from the total number of switching genes in each category. The last columns contain the percentages of genes expressed before/after the RT switch. EtoL changes = 3567 genes, and LtoE changes = 3098 genes. (B) Expression levels of the distinct classes of RT-switching genes during the EtoL (upper) and LtoE (lower) RT changes. (C) Expression levels of the distinct classes of RT-switching genes during the EtoMtoL (upper) and LtoMtoE (lower) RT changes. Transcriptional values were obtained from all respective RT changes occurring in between all intermediate stages of all differentiation pathways. D–F illustrate the transcriptional regulation and RT dynamic changes in endoderm (D), mesoderm (E), and ectoderm (F) differentiation per RT switching class. The genes with a fold difference ≥6 compared with hESC were classified into the RT clusters from Figure 4B–D and separated in the distinct categories of RT-switching genes to identify the kinetics of regulation. Line graphs depict the dynamics in RT of each cluster, and box plots were used to display the transcriptional levels across the differentiation pathways: (*) P ≤ 0.05; (**) P ≤ 0.01; (***) P ≤ 0.001 compared to hESC. Specific stages of differentiation are shown at the bottom. (G) Densities of regulatory interactions among E-class, C-class, and O-class genes. The probability of inter-class regulation was calculated by defining each class of genes by different values of RT cutoff (RTC), stringency (PCT), and switch cutoff (SC). Each PCT row is a composite of six rows representing SC values of 0.5 to 1.0 from the bottom up. Each cell represents the probability of the x-axis gene class regulating the y-axis gene class, based on the corresponding values of RTC, PCT, and SC. H-I Switching genes are preferentially located at the TTRs. Densities of the TSS from the distinct categories of genes were calculated as a function of their distance to the early replication boundary and plotted according to their RT (H) or probability densities (I). (J) Induction of C-class genes precedes EtoL RT changes and induction of E-class genes. Expression levels (top graphs) and RT changes (bottom graphs) of representative genomic regions during differentiation.











