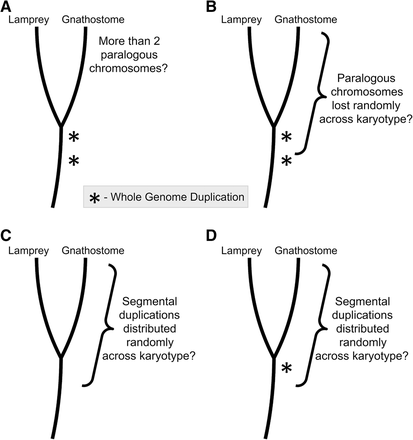
A summary of statistical tests aimed at assessing the feasibility of several evolutionary models that have been proposed to explain the distribution of paralogous regions in gnathostome genomes: (A) A liberal test of a simple model invoking two rounds of whole-genome duplication (WGD). (B–D) Tests of alternate scenarios wherein chromosome losses or segmental duplications occur in the context of a specified number of WGD events. (B) Two rounds of WGD followed by extensive loss of large duplicated segments/chromosomes. (C) A model invoking only segmental duplication. (D) A model invoking segmental duplications pre- or post-dating a single WGD. Asterisks denote whole-genome duplication events.











