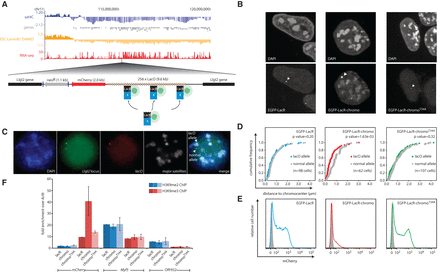
Induced proximity to chromocenters is sufficient for transcriptional repression. (A) Genomic view with sat4C, Lamin B1 DamID, and gene expression data for the lacO integration site. Below is the lacO transgene used in this assay. Note that EGFP-LacR fusion proteins are transiently transfected and recruited to the lacO array. (B) DAPI and EGFP distribution of the EGFP-LacR fusion constructs in mouse ESCs. Arrowheads highlight the position of the bright lacO integrated site. (C) Image series of three-color FISH strategy used to measure pericentromeric proximity of the lacO and untargeted allele in D. In this example, only the Llgl2 allele (green) that overlaps with the lacO transgene (red) is associated with pericentromeric satellites (white). Images are maximum projections of a z-stack to simultaneously show both the normal and lacO allele. (D) Cumulative frequency plot of the distance of the lacO (color) and normal (gray) allele to the nearest chromocenter as measured by DNA FISH as shown in C. P-values are based on two-sample Kolmogorov-Smirnov tests. Numbers of cells analyzed (with one allele each) are indicated in parentheses. (E) FACS plots of mCherry expression levels. Gray peak represents auto-fluorescence in nontransgenic founder cells. (F) ChIP data showing enrichment values of H3K9me2 (blue) and H3K9me3 (red) levels relative to LacR transduced cells. Values represent averages and error bars the standard deviations from three independent ChIPs, normalized to the Actb promoter.











