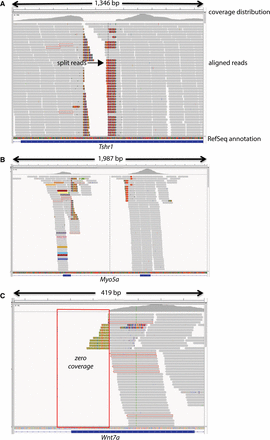
Graphical view of alignments across Tshr (A), Myo5a (B), and Wnt7a (C). Graphical views of the alignments were generated using the Integrative Genomics Viewer (IGV) and RefSeq exon annotations are shown. In each case, split reads (arrow) span the junctions of copy number variations and structural rearrangements. In Tshr, a cluster of four single nucleotide variants (SNVs) with unexpected allele frequencies of 0.3–0.63 was called in a homozygous sample; three of the four SNVs were soft filtered as a SNP cluster by GATK. Manual analysis of the alignment revealed a homozygous deletion in the final exon of this gene (A). In another example, a heterozygous SNV was called in a splice donor site of myosin VA (Myo5a) in a sample. In the alignment surrounding the SNV call there were split reads, as well as flagged reads (B, colored reads) with mates mapping throughout the genome, providing evidence of a retroviral or intra-cisternal A-particle (IAP) insertion in exon 3 (B). In a third example, a SNV call was flagged by our algorithm as a mutation candidate but could not be validated due to multiple failed PCR assays. The SNV was in wingless-related MMTV integration site 7a (Wnt7a) in an affected sample from a pedigree with recessive skeletal abnormalities. Manual analysis of the alignment surrounding the SNV call revealed two clusters of flagged reads flanking an ∼23-kb region, spanning intron 3, the 5′ splice site, and a portion of exon 3. Moreover, there was zero coverage across exon 3 and the 5′ splice site of intron 3, regions that are normally covered by WES (C).











