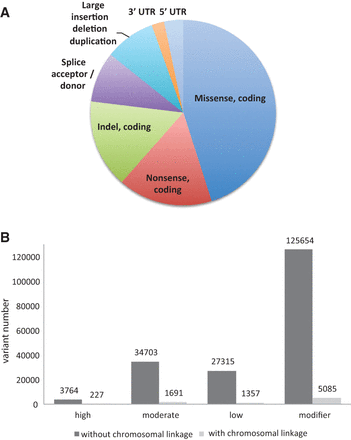
Distribution of pathogenic mutation types (A) and the value of chromosomal linkage (B) for mutation discovery in spontaneous mouse models of Mendelian disease. Pathogenic mutations consisted of a variety of lesions, the majority of which were single nucleotide substitutions. Due to ascertainment bias, copy number variants and structural mutations (>50 bp) were more rare (A). Chromosomal linkage data had a significant impact on the validation burden and a potential for false positive mutation calls. The largest effect (two orders of magnitude) was on potentially low-impact (modifier) variant calls. Variant calls were categorized by predicted impact according to SnpEff impact annotations (B) (see Methods and Supplemental File 4).











