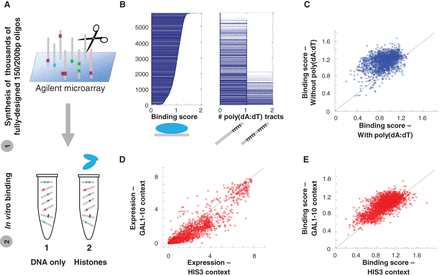
Nucleosome sequence preferences captured by BunDLE-seq suggest additional mechanisms underlying expression differences between different regulatory sequences. (A) Schematic illustration of the application of BunDLE-seq with histones. (B) Sequences were sorted according to the nucleosome binding score (computed based on the histones-bound band). (Right) For each sequence, we show the number of 15-bp poly(dA:dT) tracts that it contains. (C) Scatter plots of nucleosome binding scores for sequences with a 15-bp poly(dA:dT) tract versus the binding score of the corresponding sequences lacking this tract [among 1972 pairs tested, for 1843 the sequence without the poly(dA:dT) tracts showed a higher binding score than a corresponding sequence with the tract]. (D) Scatter plot of the expression measurements of sequences with the HIS3-derived context versus the corresponding sequences (i.e., containing the same set of regulatory elements placed at the same locations along the sequence) with the GAL1-10–derived context (with 71% below the line). (E) Scatter plots of nucleosome binding scores for the same sequences shown in D (with 80% above the line).











