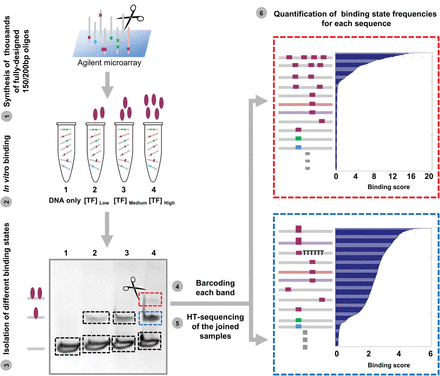
Measurements of TF binding to thousands of long, designed DNA sequences. Schematic illustration of our experimental assay, BunDLE-seq. A library of thousands of fully designed DNA sequences at lengths of 150 or 200 bp was synthesized and cleaved from Agilent programmable microarrays. These sequences differ, for instance, in their general context or their TFBS composition (with binding sites for the TF with which the experiment is carried out colored in purple, as the illustrated TF is colored, while binding sites for other TFs are colored in cyan or green). The pool of DNA sequences was incubated with buffer alone (“DNA only”) or with different concentrations of either Gcn4 or Gal4. The DNA was then run on a gel (an example of a band corresponding to DNA bound by a single TF is marked in blue, while a band corresponding to DNA bound by two TFs is marked in red), extracted from each band, amplified with a barcode marking the originating band, and sent to high-throughput sequencing. Based on the sequencing results, we computed the binding score as the ratio of the observed frequency of each sequence in each binding state (each band) versus the expected frequency (based on the “DNA only” sample). The sorted binding scores computed for a single-TF binding band (in blue) and a two-TF binding band (in red) are shown with a schematic illustration of some of the sequences that were found to be enriched in each of these bands. Filled squares represent TFBSs; filled ovals, TFs. TTTTTT represents poly(dA:dT) tracts.











