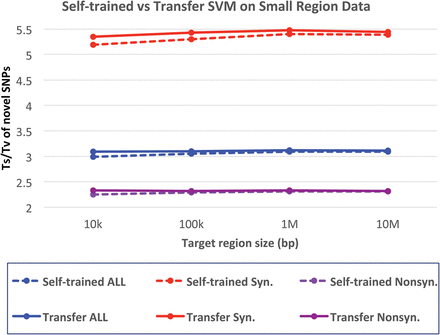
Impact of model-transfer filtering on the variant quality. The vertical axis represents Ts/Tv for novel SNPs for model-transferred SVM and self-trained SVM filters. Ts/Tv for known SNPs are ∼3.5, which is higher than novel SNPs because known SNPs contain a larger fraction of synonymous SNPs. The horizontal axis represents the size of targeted regions randomly selected from 500 whole-exome sequences. The transferred model is trained on nonoverlapping 500 exome data.











