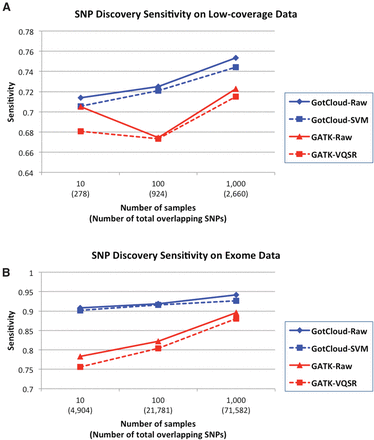
Variant discovery sensitivity comparison of GotCloud and GATK using HumanExome BeadChip excluding the SNPs contained in Omni2.5 array, because Omni2.5 variants are used to train variant filters in GotCloud and GATK. GotCloud results are shown for unfiltered (raw) and SVM-filtered sets, and GATK results are shown for unfiltered and VQSR-filtered sets, across low-coverage genome (A) and exome data (B).











