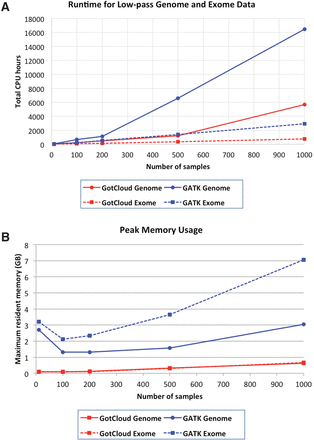
Computational costs of GATK UnifiedGenotyper and GotCloud pipelines. (A) Runtime estimated for whole-genome (6×) and whole-exome (60×) sequence data running with 40 parallel sessions on a four-node minicluster with 48 physical CPU cores. For GATK, runtimes for 1000 samples are extrapolated from analyses of a single 5-Mb block of Chromosome 20. For all other analyses, no extrapolation was used. (B) Peak memory usage estimates averaged over Chromosome 20 chunks.











