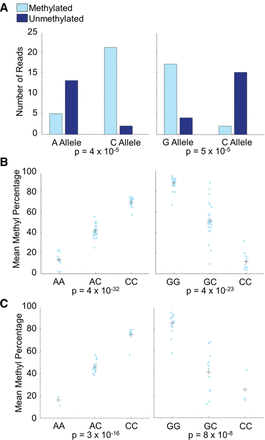
Pyrosequencing validation of mQTLs. Shown are two mQTLs involving SNPs previously identified in GWAS. (Left) The SNP is associated with age-related macular degeneration; (right) the SNP is associated with the ratio of visceral adipose tissue to subcutaneous adipose tissue. (A) Pooled bisulfite sequencing for the two mQTLs, showing strong association. (B) Pyrosequencing validation of the two mQTLs in individual samples that were used for the pooled bisulfite sequencing confirms the bisulfite sequencing results. Light blue points are the methylation percentages from individuals, and crosses are the mean methylation percentages for individuals of each genotype. (C) Pyrosequencing validation of the two mQTLs in 30 additional YRI individuals shows that the mQTLs are not limited to the individuals in our study. Light blue points are the methylation percentages from individuals, and crosses are the mean methylation percentages for individuals of each genotype.











