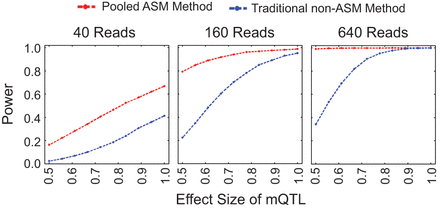
Simulations comparing our pooled ASM method to the traditional non-ASM method. Our pooled ASM method has more power to detect mQTLs than the traditional non-ASM method, especially for low effect sizes. Simulated reads from 100 individuals were sampled from a negative binomial distribution, with minor allele and minor methylation frequencies of 0.1. Power to detect an mQTL is shown for each method as a function of mQTL effect size and total read depth. The effect size is the correlation between allele and methylation status, and the power is the fraction of the simulations in which we identified an mQTL (with P < 0.001). Additional simulations are shown in Supplemental Figures 1–8, and ROC curves from the simulations are shown in Supplemental Figures 9–12.











