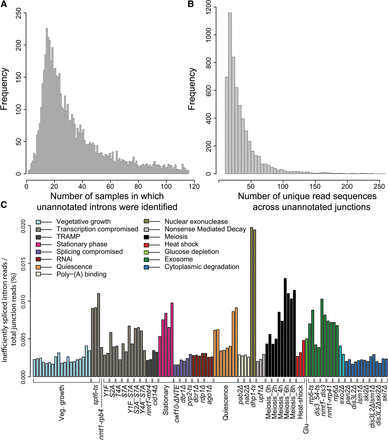
Distribution of novel introns with respect to samples in which they were identified (A) and the sum of their supporting unique read sequences across the unannotated splice junctions (B). A total of 5720 unannotated splicing events are shown. (C) Inefficiently spliced introns accumulate in nuclear surveillance and transcription mutants, meiotic differentiation, and during stationary phase and quiescence. A cutoff of 70 or fewer unique sequences across junctions was applied, resulting in ∼92% (5235/5720) of least efficiently spliced introns (for values in each sample, see Supplemental Table S12). We crudely separated potentially novel introns from cryptic introns by applying an arbitrary threshold (more than 70 unique sequences across the junction; 485/5720 putative novel introns). Ratios of sample-specific inefficiently spliced intron reads to total junction reads are shown, reflecting the proportion of exon–exon reads of inefficiently spliced introns among total exon–exon junction reads. Physiological conditions or mutants as indicated below were grouped and color coded according to cellular function or condition tested (Supplemental Table S1).











