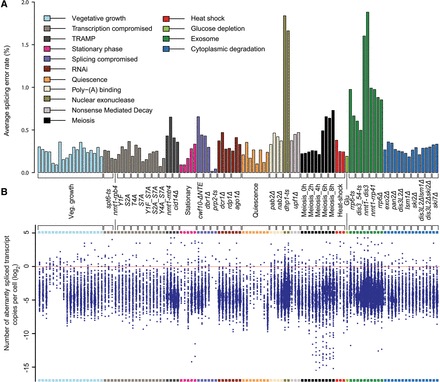
Splicing errors by exon skipping are eliminated by nuclear RNA surveillance. (A) Average splicing error for a given sample. Set of 2504 exon-skipping events for which both normal splicing and exon-skipping reads were identified in one or more samples. The splicing error rate for a locus is equal to corresponding local ESR (Supplemental Table S4). The values shown are averages of all local ESRs in a sample (Supplemental Table S7). Physiological conditions or mutants as indicated below were grouped and color coded according to cellular function or condition tested (Supplemental Table S1). (B) Number of aberrantly spliced transcripts per cell. To estimate this number, we multiplied local ESR for each exon-skipping event by corresponding transcript copy number (Supplemental Tables S8; Marguerat et al. 2012). Absolute cellular numbers of transcripts from growing cells were used as the gold standard for all samples, except for quiescence, where reported absolute numbers were used. Each blue point represents the copy number of an aberrantly spliced transcript. To avoid log2 of zero, exon-skipping events with local ESR/splicing error of zero were removed. Strains as indicated in A.











