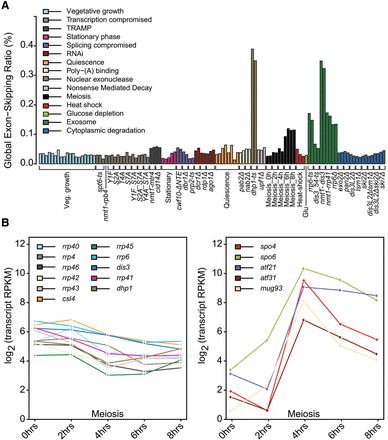
Exon-skipping events are rare but accumulate in nuclear surveillance mutants and meiosis. (A) Sample-specific global exon-skipping ratio (ESR), reflecting the proportion of exon-skipping reads among total exon–exon junction reads. Physiological conditions or mutants as indicated below were grouped and color coded according to cellular function or condition tested (Supplemental Table S1). (B) RNA-seq expression profiles of selected transcripts during meiotic differentiation. (Left) Exosome subunit and nuclear exonuclease transcripts. (Right) Selected transcripts known to increase during meiosis (Mata et al. 2002). Mean expression of two biological replicates is shown at each time point (RPKM, reads per kilobase per million). The decrease in nuclear-exosome transcripts is significant (P-adjust < 0.05, DESeq) (Anders and Huber 2010).











