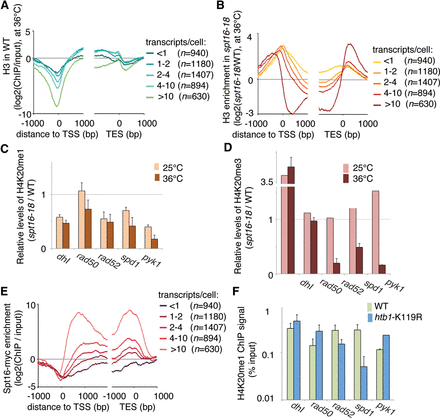
Transcription-mediated nucleosome conservation is required for the accumulation of H4K20 methylation. (A,B) Histone H3 density in WT cells (A) and changes in histone H3 density in spt16-18 cells relative to WT cells (B) and grown at permissive temperature (25°C) followed by 1 h at the restrictive temperature (36°). Data were stratified according to the average number of transcripts per cell during the vegetative growth phase. (C) ChIP-qPCR analysis of H4K20me1 at five genomic loci in spt16-18 cells and WT cells. (D) ChIP-qPCR analysis of H4K20me3 at five genomic loci in spt16-18 cells and WT cells. (E) Metagene analysis of Spt16 levels in WT cells. (F) ChIP-qPCR analysis of H4K20me1 at five genomic loci in htb1-K119R cells and WT cells. (C,D,F) Bars show the average of three independent experiments, and error bars show the SEM.











