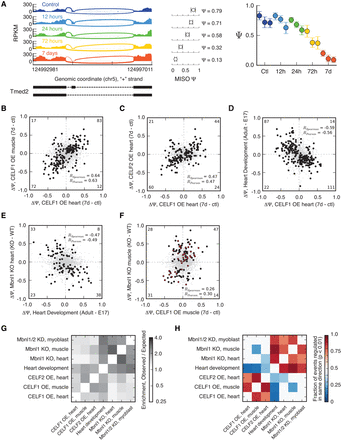
CELF1 and CELF2 regulate hundreds of splicing events, reversing many changes during heart development and antagonizing a subset of Mbnl-regulated exons. (A) RNA-seq read coverage across Tmed2 exon 3 from mouse heart at several time points following CELF1 induction. MISO Ψ values and 95% confidence intervals shown at right. (B) Splicing changes that occur following CELF1 overexpression (OE) in heart correlate with splicing changes that occur following CELF1 overexpression in muscle. n = 2496 alternative splicing events (skipped exons, alternative 3′ splice sites, alternative 5′ splice sites, retained introns, and mutually exclusive exons) shown: Monotonically changing exons shown as black circles, others as gray dots. Correlation values of monotonic events (shown) are higher than for all events: The numbers of monotonic events in each quadrant are shown in corners. (C) Splicing changes that occur in response to CELF1 overexpression in heart correlate with splicing changes that occur in response to CELF2 overexpression in heart. As in B, with n = 2129 skipped exons shown. (D) Splicing changes that occur in response to CELF1 overexpression in heart inversely correlate with splicing changes that occur during mouse heart development. As in B, with n = 1952 skipped exons shown. (E) Splicing changes that occur in Mbnl1 KO heart inversely correlate with splicing changes that occur during mouse heart development. n = 3190 skipped exons shown, as in B. (F) Splicing changes that occur in Mbnl1 KO muscle correlate with splicing changes that occur following CELF1 overexpression in muscle. n = 1501 skipped exons shown. Events that changed monotonically, or with BF > 5, following CELF1 overexpression in muscle or Mbnl1 KO in muscle, respectively, are shown in black, and correlations are listed for these events. Events that also changed monotonically during heart development are shown in red. (G) Exons regulated during heart development also tend to change in Mbnl1 and Mbnl2 knockdown myoblasts, in Mbnl1 and Mbnl2 knockout mice, and in CELF1 or CELF2 overexpressing mice. Enrichment (observed/expected number of regulated exons) is shown in the heatmap. (H) Splicing of exons regulated in response to CELF overexpression or Mbnl depletion tends to change in a direction opposite from changes that occur during heart development. The fraction of events changing in the same direction for each pair of comparisons is shown in the heatmap (only biases significant at P < 0.01 by binomial test are colored). See also Supplemental Figures S1 and S2 and Supplemental Tables S1–S4.











