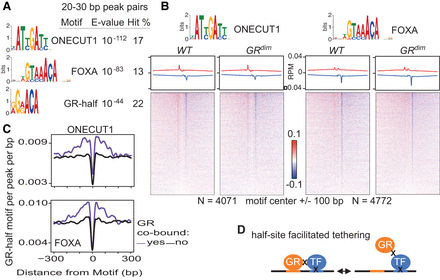
GR and GRdim occupy the liver genome at tethered sites. (A) MEME de novo sequences from 6 a.m. common site peak pairs separated by 20–30 bp and with a hit count of at least 5%. See Supplemental Material for a comprehensive list of motifs. (B) GR ChIP-exo at 6 a.m. common sites, with average profiles and density heatmaps for the ONECUT1 (left) and FOXA (right) motifs shown for both mouse models. GR common sites cobound by ONECUT1 or FOXA2 were interrogated. Red and blue indicate the 5′ ends of the forward- and reverse-stranded tags, respectively. (C) Distribution of the GR half-site motif relative to neighboring motifs at common sites cobound by ONECUT1 (top) or FOXA2 (bottom). Results for ONECUT1 and FOXA2 liver sites without GR are shown for comparison. (D) Half-site-facilitated tethering. The GR half-site motif is represented in orange. X indicates formaldehyde crosslinking between proteins or protein–DNA. Two formaldehyde crosslinking events between GR and a DNA-crosslinked TF are necessary to detect sites where GR appears bound to noncanonical motifs.











