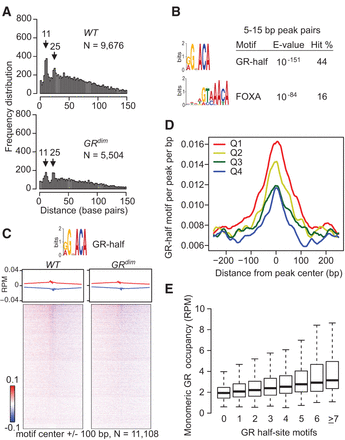
GR and GRdim occupy the liver genome at the half-site motif. (A) The distance distribution for opposite-stranded peaks with at least 0.2 RPM from GR ChIP-exo in liver isolated at 6 a.m. is shown for sites commonly bound in WT and GRdim mice, with the number of peak pairs and prominent peak distances indicated. (B) MEME de novo sequences from 6 a.m. common site peak pairs separated by 5–15 bp and with a hit count of at least 5%. See Supplemental Material for a comprehensive list of motifs. (C) GR ChIP-exo at 6 a.m. common sites, with average profiles and density heatmaps for the half-site motif shown for both mouse models. Red and blue indicate the 5′ ends of the forward- and reverse-stranded tags, respectively. (D) Distribution of the GR-half motif relative to the center of GR ChIP-seq peaks for the common sites subdivided into quartiles. (E) Plot of GR-binding strength relative to the number of half-site motifs present in ChIP-seq peaks from common sites.











