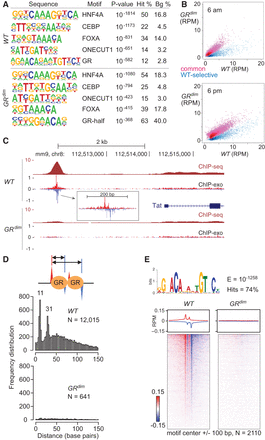
GR, but not GRdim, occupies the canonical palindromic motif as a dimer in liver. (A) Top-ranked de novo motifs from HOMER for the GR cistromes from WT and GRdim mice. See Supplemental Material for a comprehensive list of motifs. (B) Scatter plots comparing sequence tags from 14,940 GR ChIP-seq peaks with at least two reads per million (RPM) in any condition in livers isolated from WT and GRdim mice killed at either 6 a.m. (top) or 6 p.m. (bottom). Blue and red highlight WT-selective and common sites, respectively. (C) GR binding upstream of the tyrosine aminotransferase (Tat) gene. The 5′ ends of forward- and reverse-stranded sequence tags are indicated in red and blue, respectively, for the ChIP-exo tracks. Tracks are RPM normalized. (D) Distance distribution for opposite-stranded peaks with at least 0.2 RPM from GR ChIP-exo in liver isolated at 6 a.m. from WT and GRdim mice is shown for WT-selective sites. The number of peak pairs and prominent peak distances are indicated. Schematic of opposite-stranded peaks is shown at top. (E) GR ChIP-exo for WT-selective sites in liver isolated at 6 a.m. MEME top-ranked de novo sequence with a hit count of at least 5% is shown at the top. See Supplemental Material for a full list of motifs. Average profiles (middle) and density heatmaps (bottom) of the raw sequence tags are shown for both mouse models. Red and blue indicate the 5′ ends of the forward- and reverse-stranded tags, respectively.











