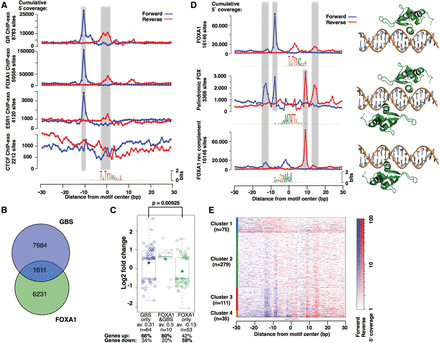
FOX motif analysis. (A) Footprint profiles for the FOXA1 motif in different ChIP-exo experiments: GR in IMR90, FOXA1 in MCF7, ESR1 in MCF7, and CTCF in HeLa cells. (B) Venn diagram showing the number of GR-bound regions with a FOXA1 and/or GBS motif in IMR90 cells. (C) Boxplot of log-fold change for genes that are differentially expressed upon treatment for 4 h with 1 μM dexamethasone (log-fold change ≤ −0.5 or ≥0.5). Genes with ChIP-seq peaks in the region ±20 kb around the TSS with only a FOXA1 motif (P < 0.0001) are marked in green; genes with peaks with only a GBS (P < 0.0001) motif in dark blue; and genes with peaks with sequences matching both motifs in turquoise. Center lines show the medians; diamonds show the mean; and box limits indicate the 25th and 75th percentiles as determined by R software; whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles. (D) FOXA1 ChIP-exo profile for the palindromic FOX motif (middle) align with those for the FOXA1 motif (top) and its reverse complement (bottom). Structural alignment indicates that the palindromic FOXA1 binding site can be simultaneously bound by two FOXK1 proteins (PDB 2C6Y), a close homolog of FOXA1. (E) K-means clustering of the 500 most occupied palindromic FOXA1 binding sites.











