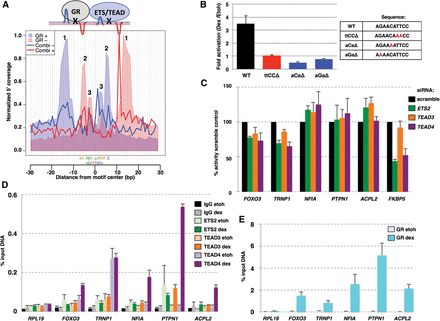
Characterization of the combi motif. (A) Alignment of the footprint profiles for the combi and the GBS motifs and a model indicating how this profile can be explained by binding of GR and another “partnering” protein. 1, “outer peaks”; 2 and 3, “inner peaks.” (B) Transcriptional activity of luciferase reporters containing a minimal promoter and three copies of the combi motif or mutant versions as indicated. Fold induction ± SEM (n ≥ 3) in U2OS cells upon treatment with 1 μM dexamethasone (dex) is shown. (C) Effect of siRNA knockdown of genes as indicated on transcriptional induction of GR target genes with nearby combi-motif-containing ChIP-seq peaks. Two days after U2OS cells were transfected with dsiRNAs, cells were treated with EtOH as vehicle control or dex, and percentage induction, relative to the scramble control, ±SEM (n ≥ 3) was determined by qPCR. (D) ETS2, TEAD3, TEAD 4, nonspecific binding (IgG) and (E) GR binding at genomic loci harboring a combi motif. ChIP experiments were performed for U2OS cells treated with EtOH as vehicle control or dex, and binding was examined for GR-bound regions with a combi motif or an unbound region (RPL19) as a control. The percentage of input immunoprecipitated ±SEM (n = 3) is shown.











