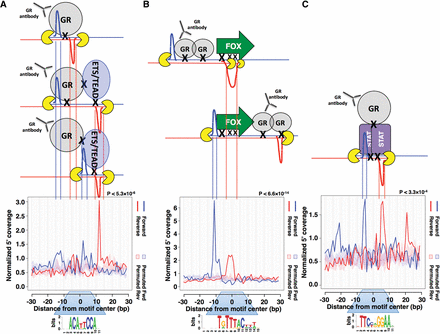
ExoProfiler identifies non-GBS footprint profiles in GR ChIP-exo data from IMR90 cells. Footprint profiles and models explaining these profiles: (A) for a de novo identified “combi” motif where monomeric GR binds together with a “partnering” protein from the ETS or TEAD family at a composite binding site; (B) for the FOXA1 motif (JASPAR MA0148.3), which could be a consequence of summarizing the reads derived from several loci at which GR and one of the FOX TFs are simultaneously cross-linked (combinatorial mode of binding); and (C) for the STAT1 motif (JASPAR MA0137.3) where GR is tethered to the DNA via STAT proteins.











