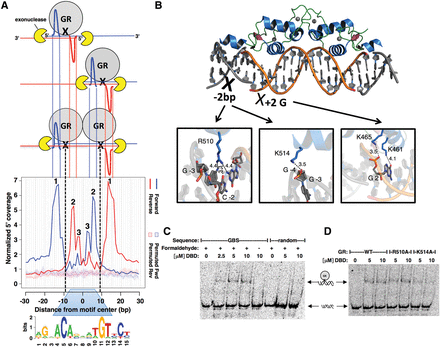
Model explaining the footprint profile for GBSs and identification of one of the GR:DNA cross-linking points. (A) Inefficiency of cross-linking and monomeric GR binding results in the cross-linking of either one or both GR monomers. Notably, a population of cells with different cross-link scenarios is analyzed, thus explaining the observed footprint profile. Dashed black lines indicate the hypothesized main DNA:GR cross-linking point (in the center of the peak-pair for each monomer). 1, “outer peaks”; 2 and 3, “inner peaks.” (B) Contacts mapping to the hypothesized GR:DNA cross-linking region based on the crystal structure of the DNA-binding domain (DBD) of GR (PDB 3G6U). (C) Denaturing EMSA identifies cross-linked DNA:GR complexes. Shifted complex is only observed on denaturing gels when DNA:GR DBD (human residues 380–540) complexes are formaldehyde cross-linked. (D) Denaturing EMSA showing that the K514A mutation results in decreased DNA:GR DBD cross-linking.











