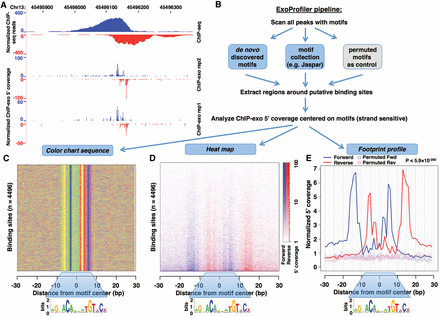
Comparison between ChIP-seq and ChIP-exo signal and a flowchart of the ExoProfiler tool. (A) GR binding coverage at the NUFIP1 locus is shown as a UCSC Genome Browser screenshot for IMR90 ChIP-seq data (top) and two ChIP-exo replicates (bottom). (B) Schematic flowchart of the ExoProfiler pipeline. The pipeline takes as input peak regions and ChIP-exo signal (mapped reads). Peak regions are scanned with motifs to find putative TFBSs and to define short regions around them; the example shown here is for a GBS motif (JASPAR MA0113.2). The 5′ ChIP-exo coverage is calculated in a strand-specific way within these regions. As output, ExoProfiler produces plots, including a color chart summarizing the sequence of motif matches (C), a heatmap displaying the 5′ ChIP-exo coverage (D), and a footprint profile recapitulating the 5′coverage for all short regions (E). As a control, this plot displays the 5′ coverage for regions matching permutated motifs. The permutations are summarized by the median (dotted line) and the interquartile range (shaded area).











