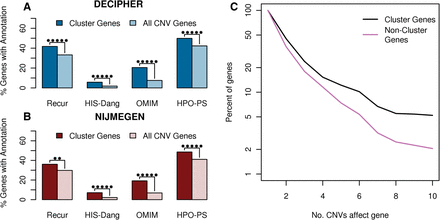
Enrichment of various disease-relevant annotations in functional clusters respectively compared to all genes in their CNVs. The enrichment of disease genes in DECIPHER (A) and NIJMEGEN (B) functional clusters. Recur indicates genes found in more than one de novo CNV in the same data set, HIS-Dang are haploinsufficient genes identified in Dang et al. (2008), OMIM are genes causally related to a disease in the OMIM database (OMIM 2012) and HPO-PS are candidate genes specifically associated with the respective patient's phenotype based on gene-phenotype annotations in the Human Phenotype Ontology database (Dolken et al. 2012). Stars indicate significance: (*) P < 0.05, (**) P < 0.005, (***) P < 0.0005, etc. up to a maximum of five stars. (C) Survivorship curve indicating the frequency of functional cluster genes in recurrent regions compared to CNV genes not belonging to clusters of functionally related genes. The more frequently a gene was seen affected by de novo CNVs, the greater the chance it belongs to a functional cluster.











