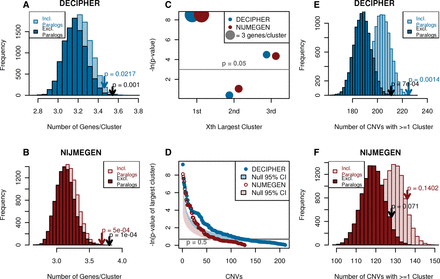
De novo CNVs from patients with developmental disorders contain significantly large numbers of functionally similar genes, as defined by proximity in the phenotypic linkage network (PLN). (Blue) DECIPHER, (red) NIJMEGEN. (A,B) DECIPHER (A) and NIJMEGEN (B) de novo CNVs contain significantly large functional clusters compared to 10,000 gene-number-matched randomizations, the significance of which increases when paralogous genes within the same CNV are collapsed to a single copy. Arrows indicate observed value and P-value. (C) The largest functional cluster is most significant in both data sets. The size of the circle indicates the average cluster size, light gray line indicates P = 0.05, and data sets are offset due to high overlap. (D) Thirty percent of de novo CNVs contain a functional cluster that is larger than expected (points, gray line indicate P = 0.5); shaded areas indicate 95% confidence intervals given a uniform distribution of P-values. The respective patients were not significantly enriched for any phenotype (hypergeometric test with Bonferroni correction). (E,F) More DECIPHER (E) and NIJMEGEN (F) de novo CNVs contain functional clusters compared to 10,000 gene-number-matched randomizations. Only DECIPHER was significantly different. Arrows indicate observed value and P-value.











