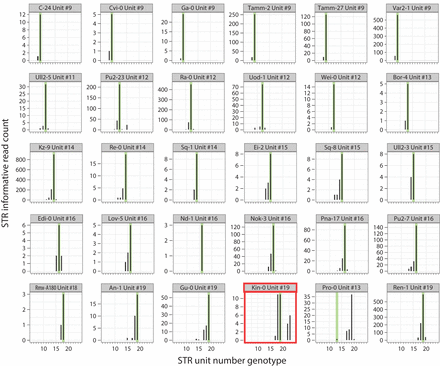
MIPSTR accurately determined germline ELF3 STR unit number across genetically diverse A. thaliana strains. Histograms of raw read counts across 30 accessions. STR unit number as determined by Sanger sequencing is indicated in green. Using tag-defined read groups, the Kin-0 ELF3 STR genotype can be resolved to the known STR genotype even with comparatively few total reads. MIPSTR clearly calls STR unit number 19 for Pro-0. Note that different individuals of the same strain were analyzed with MIPSTR and Sanger sequencing, which may explain the discrepancy. Bold red outline indicates example discussed further in the text.











