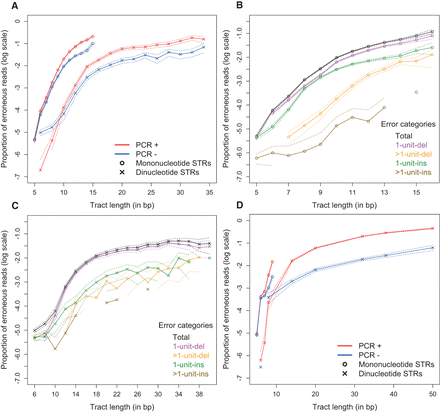
Erroneous call rates for Illumina data. The dotted lines represent the 95% confidence intervals of the multinomial sampling. Only repeat lengths with ≥100 read support (all loci combined) were plotted. (A) Human male X Chromosome for PCR-containing (PCR+) and PCR-free (PCR−; down-sampled data) library preparation protocols. (B) PCR− mononucleotide STR error rates by error categories. See text for the explanation of the categories. (C) PCR− dinucleotide STR error rates by error categories. (D) Error rates for ultradeep sequencing of plasmids with PCR+ and PCR− protocols.











