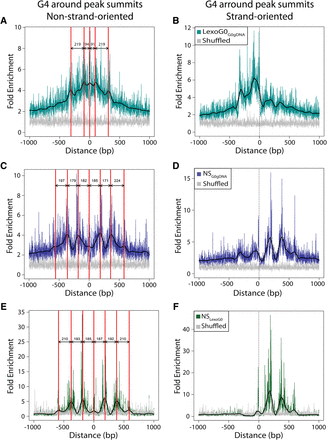
Distribution of G4 motifs around peak summits. (Left) The distributions of G4 motifs (non-strand-oriented) around LexoG0G0gDNA (A), NSG0gDNA (C), and NSLexoG0 (E) peak summits. The red lines indicate the positions of wave crests; labeled arrows, the distance between adjacent crests. (Right) The distributions of G4 motifs (strand oriented 5′-3′ left to right) around LexoG0G0gDNA (B), NSG0gDNA (D), and NSLexoG0 (F) peak summits.











