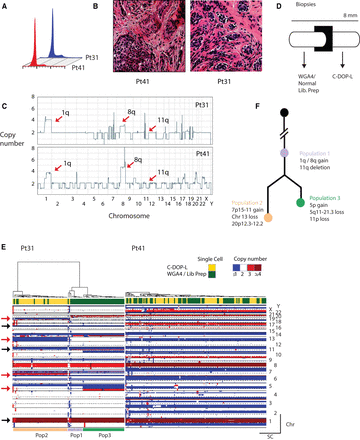
Highly multiplex single-cell sequencing identifies genomic heterogeneity in breast cancer biopsies and informs evolutionary history. (A) Flow cytometric profiles of the clinical cases. (B) Similar histopathologies of the biopsies showing invasive ductal carcinoma with moderate differentiation and complex glandular growth pattern. (C) Bulk genome-wide copy number profile of the two cases. Arrows point to genomic regions recurrently gained or lost in ER + breast cancer. (D) Schematic representation of biopsy sectioning and processing using single-cell methods. (E) Hierarchical clustering heatmap of the Pt41 and Pt31. Arrows point to genomic regions shared by both tumor genomes (black) and genomic regions that display heterogeneity in Pt31 (red). (F) Evolutionary history of Pt31 subpopulations.











