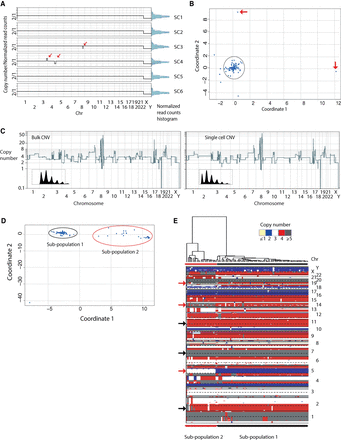
C-DOP-L approach provides uniform, unbiased amplification of single-cell genomes, accurate determination of copy number states and reveals genomic heterogeneity in breast cancer cell lines. (A) Genome-wide copy number plots of single cells at 5K bins of a diploid karyotypically normal lymphoblastoid cell line from a male (315A) and histogram distributions of normalized read count data. Red arrows point to examples of somatically mosaic integer copy number events that are observed in some single cells. Data plotted are of nonrounded copy number estimates. (B) Multi-dimensional scaling of 95 single cells from 315A. Red arrows point to outlier cells. (C) Copy number profile of a single SK-BR-3 cell (right) compared to bulk from millions of cells (left) at 5K bins. Boxed inserts display smoothened kernel density plots of normalized read counts showing discrete densities (quantal data). (D) Multidimensional scaling of 94 single SK-BR-3 cells. (E) Hierarchical clustering copy number heatmap of single-cell SK-BR-3 genomes. Black arrows denote copy number alterations shared by the vast majority of single cells. Red arrows denote copy number alterations that distinguish the two subpopulations.











