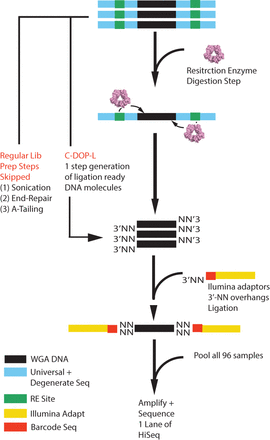
Schematic overview of the C-DOP-L approach for highly multiplex single-cell sequencing. In brief, WGA DNA is treated with restriction enzyme to cleave the universal sequences found at the ends of WGA DNA. The digestion reaction leaves 3′-NN overhangs (where N is any base: A,T,C,G). Digested DNA is then ligated to barcoded Illumina sequencing adaptors that are designed to contain 3′-NN overhangs. After barcode addition, samples are pooled, amplified, and sequenced on a single lane of the HiSeq instrument. (WGA) Whole-genome amplified, (RE) restriction enzyme, (N) any base (A,T,C, or G).











