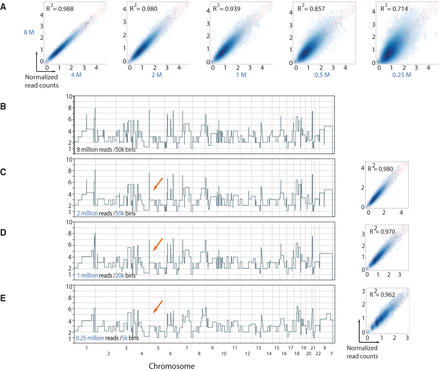
Down-sampling analysis reveals minimal data requirements for copy number determination. (A) Density scatter correlation plots of the normalized bin read counts (directly proportional to copy number) of the original 8 million uniquely mapped read data set with data sets down-sampled to 4, 2, 1, 0.5, and 0.25 million reads using 50K bins. (B) Genome-wide CNV plot of rearranged cancer cell with 8 million reads using 50K bins. (C) Genome-wide CNV view of same cell with data sets down-sampled to 2 million reads using 50K bins. Box plot illustrates normalized read count scatter correlation plots with original 8 million read data set. (D,E) Same as in C but with 1 million and 0.25 million reads using 20K and 5K bins, respectively. Red arrows exemplify CNVs that are lost with decreasing resolution (i.e., fewer number of bins). Pearson's correlation coefficients of data sets are displayed.











