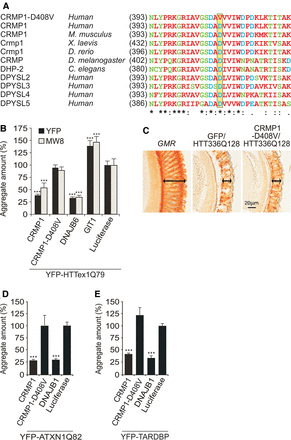
A mutant variant of CRMP1 (D408V) shows impaired activity in cell-based assays and HD transgenic flies. (A) Multiple sequence alignment of CRMP1 protein sequences. The aspartic acid residue 408 in human CRMP1 is conserved in a large number of related proteins (yellow box). (B) Effects of mCherry-tagged CRMP1 and CRMP1-D408V fusion proteins on aggregation of YFP-tagged HTTex1Q79 in cell-based assays. Formation of insoluble YFP-HTTex1Q79 aggregates in SH-EP cells was quantified by high-content fluorescence imaging. Aggregates were detected by YFP fluorescence of HTT aggregates or by immunostaining using the anti-HTT antibody MW8. MW8 preferentially detects insoluble HTT aggregates. Data are represented as mean ± SEM. (***) P ≤ 0.001, two-sided t-test with unequal variance. (C) Retinal degeneration in control flies carrying the GMR-GAL4 driver (GMR) alone and in flies coexpressing HTT336Q128 with CRMP1-D408V or the GFP control protein under control of the GMR driver. Analysis of eye sections reveals that in contrast to wild-type CRMP1, neither GFP nor CRMP1-D408V affects HTT336Q128-induced retinal degeneration compared to the GMR flies. (D–E) Effects of mCherry-tagged CRMP1 or CRMP1-D408V on aggregation of YFP-tagged ATXN1Q82 (D) or TARDBP (E) in cell-based assays. (D) YFP-ATXN1Q82 or (E) YFP-TARDBP aggregates were detected in SH-EP cells by YFP fluorescence and quantified by high-content fluorescence imaging. Data are represented as mean ± SEM. (***) P ≤ 0.001, two-sided t-test with unequal variance.











