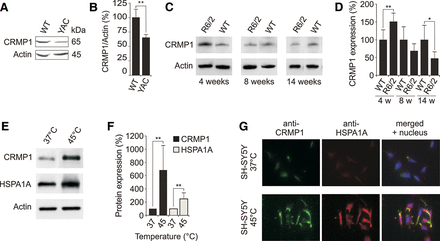
Expression of CRMP1 in brains of HD transgenic mice and cell models. (A) Endogenous CRMP1 protein levels are reduced in brain tissues of 12-mo-old YAC128 HD transgenic mice compared to controls. Protein extracts prepared from striatal tissues of wild-type and transgenic HD mice were analyzed by SDS-PAGE and immunoblotting using the anti-CRMP1 antibody 504-518. (B) Quantification of ∼65 kDa CRMP1 bands in A. Values represent the means of three independent experiments. (**) P ≤ 0.01, two-sided t-test with unequal variance. (C) Endogenous CRMP1 levels are altered in brains of R6/2 HD transgenic mice compared to controls. Protein extracts were analyzed by SDS-PAGE and immunoblotting using the anti-CRMP1 antibody 504-518. (D) Quantification of CRMP1 protein bands shown in C. Values represent the means of three independent experiments. (*) P < 0.1; (**) P ≤ 0.01, two-sided t-test with unequal variance. (E) Heat stress induces endogenous CRMP1 protein levels in SH-SY5Y neuroblastoma cells. Cell lysates were analyzed by SDS-PAGE and immunoblotting. (F) Quantification of CRMP1 and HSPA1A protein bands shown in E. Values represent the means of three independent experiments. (**) P ≤ 0.01, two-sided t-test with unequal variance. (G) Analysis of CRMP1 and HSPA1A protein expression in SH-SY5Y cells under heat stress conditions by immunofluorescence microscopy. Data in B, D, and F are represented as mean ± SEM.











