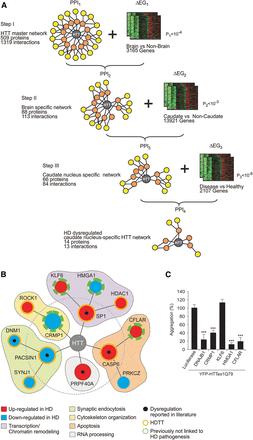
Predicting caudate nucleus-specific, dysregulated HTT-associated proteins by interaction network filtering. (A) Data integration strategy for interaction network filtering by differentially expressed genes. By systematic integration (three filtering steps) of protein interaction and gene expression data (PPI1-3 and ΔEG1-3), a caudate nucleus-specific HD network (PPI4) with potentially dysregulated, disease-relevant HTT-associated proteins was created. (B) Potential function of HTT-associated proteins predicted though systematic HTT interaction network filtering. In PPI4, 13 potentially dysregulated proteins are directly or indirectly linked to HTT. The orange ring indicates known HD therapy targets (HDTTs). (C) Effects of the computationally predicted proteins CRMP1, KLF6, HMGA1, and CFLAR on mutant HTT aggregation in cell-based assays. Potential modulator proteins were coproduced as mCherry-tagged fusions with YFP-tagged HTTex1Q79 fusion protein in SH-EP neuroblastoma cells. Formation of YFP-HTTex1Q79 aggregates was quantified after 48 h by high-content fluorescence imaging. Data are represented as mean ± SEM. (***) P ≤ 0.001, two-sided t-test with unequal variance.











