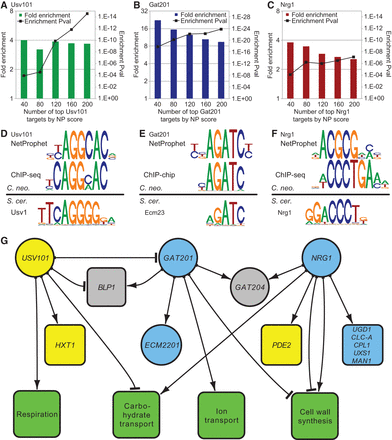
Validation of the NetProphet network. (A–C) ChIP enrichment of NetProphet predicted targets: (colored bars) cumulative fold-enrichment of the top NetProphet-predicted targets of each TF for ChIP-positive targets, relative to the fraction of ChIP-positive targets among all genes; (square symbols) significance of the enrichment (pval). The horizontal axis indicates the number of top-ranked NetProphet-predicted targets considered. (D–F) Binding motifs. For the indicated TFs, C. neoformans binding motifs derived from the promoters of NetProphet-predicted targets (top) or from the regions around ChIP-seq peaks (middle) are compared to S. cerevisiae binding motifs (bottom). (G) Network diagram showing the three validated TFs with targets supported by both NetProphet and ChIP analysis that are relevant to cryptococcal virulence: (round nodes) TFs; (square nodes) target genes or gene ontology biological process terms for which the targets of the indicated TF are enriched; (blue nodes) mutants are hypocapsular; (yellow nodes) mutants are hypercapsular; (gray nodes) mutants are defective in capsule-independent phagocytosis. Edges with arrowheads indicate activation, whereas those with T-heads indicate repression. ChIP evidence suggests that Usv101 binds to its own promoter, but expression evidence cannot determine whether this binding results in activation, repression, or no effect.











