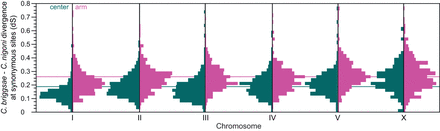
Divergence at synonymous sites for orthologs of C. briggsae and C. nigoni is higher for genes linked to arm domains than center domains for all chromosomes (all Wilcoxon test P ≤ 0.0035). The chromosomes also differ significantly from one another in average substitution rates (F(5,5085) = 72.0; P < 0.0001), with Chromosome I being lowest and Chromosomes V and X being highest (Tukey's range test). Horizontal lines indicate mean dS for genes in center domains (cyan, dS = 0.186) or arms (magenta, dS = 0.259) across all chromosomes. Loci with strong codon bias (ENC < 45) excluded from analysis.











