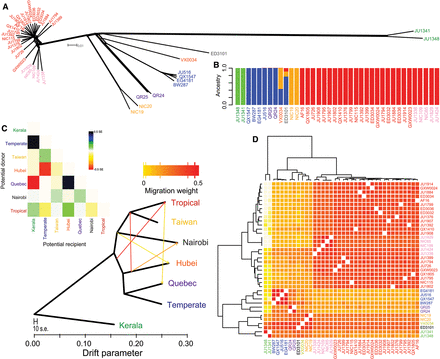
Diverse genomic analyses affirm the genetic distinctiveness of phylogeographic groups within C. briggsae. A neighbor network for all chromosomes (A) discriminates phylogeographic groups of strains, corresponding to the pan-global “Tropical” clade (red and pink strain labels), pan-global “Temperate” clade (blue), and genomic haplotype groups that exhibit restricted geographic origins around the globe: (Quebec) purple; (Nairobi) black; (Hubei) orange; (Taiwan) yellow; (Kerala) green. (B) The ADMIXTURE program minimizes the cross-validation error of ancestral relationships when it identifies four genetic clusters in this data set. (C) Permitting migration in the genomic ancestry of the 37 C. briggsae strains with TreeMix suggests multiple plausible instances of migration, although incomplete sorting of ancestral polymorphism provides an alternate interpretation. Heatmap above the genealogy indicates residual fit to a model with five migration events. (D) Haplotype clustering of the phylogeographic groups is recapitulated in a similar manner in ChromoPainter's genome-wide coancestry matrix (Euclidean log2). Dendrogram on left indicates strain clustering with the unlinked model; top dendrogram indicates strain clustering with the linked model. All analyses used the set of 439,139 SNPs with allele information present in all strains.











