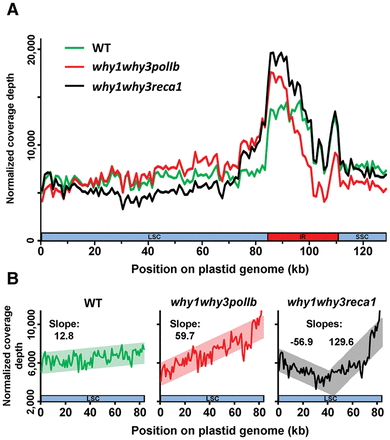
Plastid DNA sequencing coverage curves for Arabidopsis lines WT, why1why3polIb, and why1why3reca1. (A) Plastid sequencing coverage of pools of 14-d-old Arabidopsis seedlings of the indicated genotypes. Positions were rounded down to 1 kb. All reads mapping to the plastid large inverted repeats (IRs) were only assigned to the first IR. The plastid large single-copy region (LSC), the first IR, and the small-single copy region (SSC) are depicted as a long blue bar, a red bar, and a short blue bar, respectively. The y-axis represents the number of reads per 1,000,000 total plastid reads. (B) Regression analysis of the plastid LSC sequencing coverage of WT, why1why3polIb, why1why3reca1 seedlings. Positions were rounded down to 1 kb. The y-axis represents the number of reads per 1,000,000 total plastid reads.











