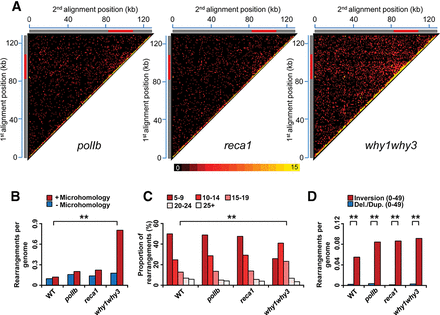
Global portrait of plastid genome rearrangements in Arabidopsis mutant lines polIb, reca1, and why1why3. (A) Plastid rearrangement breakpoint positions of the indicated Arabidopsis mutant lines. Heatmaps depict each rearrangement as the intersection of the two genomic positions corresponding to the nucleotide on each side of the junction. Each tile represents a region spanning 1 kb along each axis. Tile intensity represents the number of rearrangements per 10,000 plastid genomes. All rearrangements mapping to the plastid large inverted repeats (IRs) were only assigned to the first IR. The plastid large single-copy region (LSC), the first IR, and the small-single copy region (SSC) are depicted as a long gray bar, a red bar, and a short gray bar, respectively. (B) Normalized amount of rearrangements per plastid genome displaying a microhomology (≥5 bp) (red) or not displaying a microhomology (blue) at their junction for each of the indicated mutant lines. (C) Proportion of total plastid rearrangements displaying a microhomology of given lengths, in bases, in each mutant line. The y-axis represents the percentage of usage of each microhomology length. (D) Normalized amount of rearrangements corresponding to short-range inversions (<50 bp, U-turn-like rearrangements, red) and short-range deletions/duplications (<50 bp, blue) in each mutant line. Data were normalized on the total number of plastid genome reads. (Del.) deletion, (dup.) duplication. Two asterisks indicate a significant difference with P-value < 0.00001 using a χ2 test.











