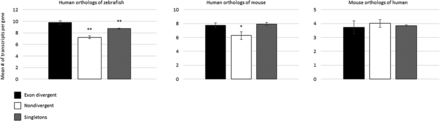
The potential for a preduplication bias. Histograms comparing the levels of alternative splicing in single-copy orthologs that serve as a proxy for the ancestral state. (Left) Human orthologs of zebrafish exon divergent paralogs (black) display higher levels of alternative splicing compared to the human orthologs of zebrafish nondivergent paralogs (white) and zebrafish singletons (gray). (Middle) Human orthologs of mouse exon divergent paralogs (black), nondivergent paralogs (white), and singletons (gray). (Right) Mouse orthologs of human exon divergent paralogs (black), nondivergent paralogs (white), and singletons (gray). The asterisks indicate the significance of the difference as compared to the singleton orthologs of the exon divergent paralogs: (**) P < 0.01 (*) P < 0.05. Error bars, SEM.











