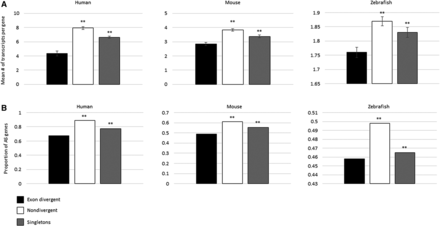
The relationship between exon structure divergence among paralogs and levels of alternative splicing. The exon divergent paralogs demonstrate significantly lower levels of alternative splicing compared to nondivergent paralogs and singletons as measured by mean number of transcripts (A) and the proportion of genes that are alternatively spliced (AS) (B). Student's t-test was used to calculate the significance of the differences in mean number of transcripts. χ2 was used to calculate the significance of the differences in proportion of genes that are alternatively spliced. The asterisks indicate the significance of the difference as compared to exon divergent paralogs: (**) P < 0.01. Error bars, SEM.











