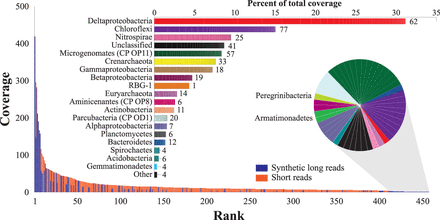
Rank abundance curve for the 5-m community including all species for which the rpS3 gene could be recovered. (Bottom) While most of the rpS3 genes were recovered from the short-read assembly (orange), least and most abundant species were represented almost exclusively by the long-read data (blue). (Top) Stacked bar graph shows abundance of phyla and Proteobacteria classes; stacked boxes indicate abundance of individual species (number of species indicated). Deltaproteobacteria is the most abundant lineage in the sample, with five of the seven most abundant species being closely related. (Pie chart) Species with zero short-read coverage in the short-read data, detected in the synthetic long reads only.











