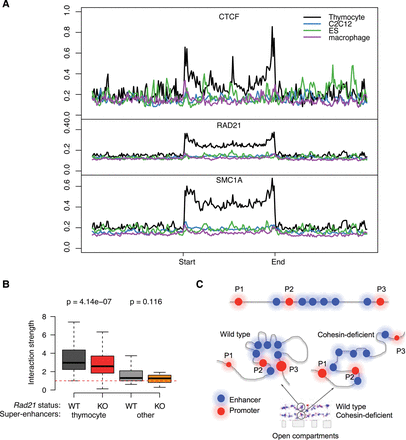
Cell-type specificity of CTCF association and spatial clustering of super-enhancers. (A) CTCF (top), RAD21 (middle), and SMC1A (bottom) ChIP-seq signal in thymocytes at super-enhancers that are active in thymocytes (black), macrophages (purple), myoblasts (blue), or ES cells (green). (B) Preferential interactions within cell-type-specific super-enhancers. SIMA analysis of thymocyte Hi-C data was used to compare interactions within super-enhancers active in thymocytes or other cell types. (Left) Hi-C interactions in control (gray) and Rad21-deficient thymocytes (red) within thymocyte super-enhancers sized 50 kb or more (n = 105). (Right) Hi-C interactions in WT (light gray) and Rad21-deficient thymocytes (orange) within super-enhancers of 50 kb or more that are active in pro-B cells, macrophages, or ES cells (n = 20) (Whyte et al. 2013). P-values shown are from a Wilcoxon signed-rank test. (C) Model for the impact of cohesin-dependent enhancer-enhancer interactions on gene expression. Spatial clustering between enhancer elements can affect promoter activity positively and negatively. The promoters P1 and P3 are both distal to a super-enhancer. P1 is isolated as a result of the spatial clustering between the enhancer elements within the SE, while P3 is connected to the super-enhancer by enhancer-enhancer contacts (left). Removal of cohesin (right) decreases the spatial constraint on enhancer elements so that P1 is contacted more readily by enhancer elements, while P3 dissociates from the super-enhancer.











